Visualization of unstained DNA nanostructures at 20 kV
May 10, 2019 - TEM visualization of individual DNA molecules in their native unlabeled form has remained extremely challenging. A team of scientists now compared different strategies to obtain high contrast without elaborate sample preparation: Cs/Cc corrected microscopy at very low primary energies, volta-potential phase plate microscopy, and dark-field microscopy choosing the easiest route of sample preparation, which is supported on commercially-available carbon membranes. SALVE data acquisition and processing allowed reconstruction of the DNA using no more than 100 room temperature micrographs.
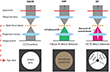
The authors compared three major electron microscopy techniques for single particle analysis (SPA) of a DNA structure. SPA, combines several digitized transmission electron microscopy (TEM)images of similar particles together, is a major image processing technique for the analysis of biological specimen. It gives an image with stronger and more easily interpretable features. The study demonstrated that imaging is not well possible with defocused CTEM at 200-300 kV. Low-kV (80 kV) non-corrected CTEM facilitates the DNA analysis but still provides insufficient resolution. Sub-Ångstrom low voltage electron microscopy (SALVE) with Cc + Cs correction at 20 kV strongly improves the contrast and the resolution. Similarly, a volta-potential phase plate can be used to boosts the DNA contrast at high voltages substantially. Dark-field microscopy could provide the necessary contrast for DNA visualization at high voltages but is presently incompatible with SPA due to limitation in the software and automation of the available instrumentation.
"The idea was to discuss the advantages and drawbacks of these techniques for broad applications in structural biology and materials science," Kaiser said. "Despite the importance of DNA in all forms of life, TEM visualization of individual DNA molecules in its native unlabeled form would provide a promising approach for biophysical assays such as probing sequence-specific protein interactions."
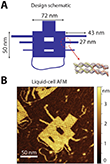
In their new work, the scientists from the Kavli institute in Delft and the Maastricht Multimodal Molecular Imaging Institute in the Netherlands, and the Group of Electron Microscopy for Materials Science at Ulm University in Germany aimed to image unstained DNA nanostructures without elaborate sample preparation techniques. They used commercially-available carbon membranes which are accessible for every TEM lab. For reliable evaluation of the image contrast, they utilized single-layer DNA „origami“ nanostructure (Fig. 1). It enables the application of single particle analysis (SPA) to a purely nucleic acid structure, instead of, for example, using DNA-bound protein structures.
Until now, unstained DNA had only been made visible if freely suspended, i.e., without carbon support for a limited number of specific DNA structures such as DNA bundles or fibers [1-6] When DNA is deposited onto commercial carbon membranes in dry condition, good contrast could not be achieved until now. Similarly, reducing the carbon thickness to increase the DNA contrast was found to be non trivial due to difficulties in manufacturing and handling of delicate carbon membranes, and because amorphous carbon below 4 nm thickness[7] might be less conductive, which strongly deteriorates the TEM imaging.
Electron optics in TEM have seen multiple improvements in recent years. However, while the conventional TEM (CTEM) is an ideal tool for imaging materials science samples with atomic resolution, the visualization of radiation-sensitive biological molecules such as DNA is an ongoing challenge. Recently, electron microscopists have developed phase contrast techniques under low-dose conditions with the help of phase plates and low-voltage TEM. For instance, a commercial phase plate is available since 2015, since Previous generations of phase plates including the well-known Zernike-type were difficult to apply for the SPA application[11] And chromatic aberration (Cc) at very low accelerating voltages of 20 kV was realized very recently starting from 2017, when the SALVE (sub-Ångstrom low voltage electron) microscope was finalized[9]. Now, the scientists presented different approaches by modification TEM of electron optics as shown in Fig. 2.
Extending TEM application to life sciences
The authors found, that the DNA origami reconstruction using the low-kV dataset at room temperature and VPP dataset at cryogenic temperature provide excellent resolution. The detailed DNA structural features such as the smaller cavity and the single DNA helices become visible. Increased total scattering cross section at 20 kV, together with enhanced low-frequency information transfer, boosts the phase contrast of the DNA.
By presenting first images of single-helix-thick unstained DNA nanostructures supported on commercial carbon supports at 20 kV the new study signifies a significant progress in biomolecular imaging. This enabled the particle picking and class averaging algorithms in SPA. The SALVE images of the DNA origami extend its application to life science specimens.
Resource: Kabiri, Y., Ravelli, R.B.G., Lehnert, T., Qi, H., Katan, A. J., Roest, N., Kaiser, U., Dekker, C., Peters, P. J. & Zandbergen, H. (2019) Visualization of unstained DNA nanostructures with advanced in-focus phase contrast TEM techniques. Scientific reports, 9, 7218, doi: 10.1038/s41598-019-43687-5.
-
Marini, M. et al. The structure of DNA by direct imaging. Sci. Adv. 1, 1–5 (2015).
-
Gentile, F. et al. Direct imaging of DNA fibers: The visage of double helix. Nano Lett. 12, 6453–6458 (2012).
-
Takai, Y., Nomaguchi, T., Matsushita, S. & Kimura, Y. Molecular-scale imaging of unstained deoxyribonucleic acid fibers by phase transmission electron microscopy. Appl. Phys. Lett. 89, 133903 (2006).
-
Company, N. P. Direct imaging of a double-strand dna molecule. Ultramicroscopy 7, 189–192 (1981).
-
Marini, M. et al. Imaging and structural studies of DNA-protein complexes and membrane ion channels. Nanoscale 9, 2768–2777 (2017).
-
Martin, T. G. et al. Design of a molecular support for cryo-EM structure determination. Proc Natl Acad Sci 7, 7456–7463 (2016).
-
Larson, D. M., Downing, K. H. & Glaeser, R. M. The surface of evaporated carbon films is an insulating, high-bandgap material. J. Struct. Biol. 174, 420–423 (2011).
-
Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA 111, 15635–40 (2014).
-
Linck, M. et al. Chromatic Aberration Correction for Atomic Resolution TEM Imaging from 20 to 80 kV. Phys. Rev. Lett. 117, 1–5 (2016).
-
Zhang, C., Xu, Q., Peters, P. J. & Zandbergen, H. The use of a central beam stop for contrast enhancement in TEM imaging. Ultramicroscopy 134, 200–206 (2013).
-
Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA 111, 15635–40 (2014).
-
Kabiri, Y., Ravelli, R.B.G., Lehnert, T. et al. Visualization of unstained DNA nanostructures with advanced in-focus phase contrast TEM techniques. Sci Rep 9, 7218 (2019) doi:10.1038/s41598-019-43687-5
-
Glaeser, R. M. Invited Review Article: Methods for imaging weak-phase objects in electron microscopy. Rev. Sci. Instrum. 84, 1–17 (2013).